Learn Python
Learn Data Structure & Algorithm
Learn Numpy
Pandas Introduction
Pandas Series
Pandas DataFrame
Pandas Read Files
Pandas Some functions and properties
Pandas Math Function
Pandas Selection
Pandas Change Type
Pandas Concatenate & Split
Pandas Sorting
Pandas Filter
Pandas Data Cleaning
Pandas Group by
Pandas Time Series
sPandas Analysis 1
Pandas Analysis 2
Pandas Analysis 3
Matplotlib
Learn Seaborn
Learn Statistics
Learn Math
Learn MATLAB
Learn Machine learning
Learn Github
Learn OpenCV
Learn Deep Learning
Learn MySQL
Learn MongoDB
Learn Web scraping
Learn Excel
Learn Power BI
Learn Tableau
Learn Docker
Learn Hadoop
data cleaning methods using pandas
Different types of missing data
Missing completely at random:
We called a variable missing completely at random when there is no relationship between the missing data and
any other values or with the dataset. We can also say that all those data points are a random subset of the
data.
Missing data not at random:
We called a variable missing data not at random when there is absolutely some relationship between the missing
data and any other values or with the dataset. We can also say that all those data points are not a random
subset of the data.
Ways to handle the missing values:
Drop the row:
This means, suppose a row containing missing values. In this method, we will ignore(delete) that row that
contains missing values.
We do this when the row doesn't that much important.
But if the row is important then we never use this method.
So we can say that we use this method on low priority or less important rows.
Drop the column:
If a column contains too many missing values then we drop that column.
Use global constants:
This means we will fill all the missing using one value like null, missing, sorry, ignore, etc.
Because we are filling all the missing values using one value so that we call that value global constant
But we don't use this technique. Because this can be the reason for bad accuracy.
Fill missing value using mean, median or mode:
Suppose a row has a missing value and the missing value column name is price.
Now we will take the mean or median or mode of that column and will fill that column with all missing cells
using that mean or median or mode value.
For categorical column, we can only use mode
For numerical column we can use mean, median or mode
Now there can be a question about when we should use mean and median?
The answer is when the data is normally distributed then we will use mean
and if the data is skewed distributed then we will use median.
Use of algorithms:
In this method, we use machine learning algorithms to predict the value of the missing cells. This an advance
technique.
Note:
Look it doesn't matter which technique we are using but the main thing we do is that we fill the missing
values or delete the missing values.
Here we select a range. If the row or column contains more missing values from the range then we drop the row
or column and if contains less from the range then we try to fill the missing value.
When we delete the missing value?
Here we drop the row or column which contains missing values.
Suppose there is a column that contains 80% missing values. In this case, we will delete that column.
Suppose there are 1 million data and only 100 data are missing. In this case most of the time we delete the
missing value.
When do we fill the missing value?
Now suppose a column contain 10% missing value.
In this case if the data is important then we fill the missing value.
Now suppose a column contains 5% missing value.
In this case, we will see the number of data and the importance of that data. If the data is very important
then we try to fill the missing values and if the data is not that important then we delete the cells.
Let's see the dataset
import pandas as np
df=pd.read_csv("practice2.csv")
print(df)
Output
| total_bill | sex | time | tip | size | day | smoker | |
|---|---|---|---|---|---|---|---|
| 0 | 500.0 | male | lunch | 30 | 2 | sat | no |
| 1 | 648.0 | male | dinner | 35 | 3 | sat | yes |
| 2 | 75.0 | female | NaN | 10 | 1 | sat | no |
| 3 | 159.0 | female | NaN | 12 | 4 | sat | NaN |
| 4 | 250.0 | male | lunch | 3 | 3 | NaN | no |
| 5 | 222.0 | female | lunch | 25 | 3 | mon | yes |
| 6 | 356.0 | male | dinner | 30 | 4 | mon | NaN |
| 7 | 99.0 | female | NaN | 5 | 1 | mon | no |
| 8 | 150.0 | male | dinner | NaN | 4 | mon | NaN |
| 9 | NaN | female | lunch | 38 | 4 | mon | no |
| 10 | 478.0 | female | lunch | 28 | 3 | tue | yes |
| 11 | 320.0 | nan | NaN | 14 | 3 | tue | yes |
| 12 | NaN | male | lunch | 32 | 2 | tue | NaN |
| 13 | 520.0 | male | lunch | 33 | 3 | NaN | yes |
| 14 | 367.0 | male | dinner | NaN | 3 | tue | yes |
| 15 | NaN | female | NaN | 40 | 4 | wed | yes |
| 16 | 100.0 | female | dinner | 6 | 1 | wed | NaN |
| 17 | 99.0 | Nan | NaN | 40 | 1 | wed | yes |
| 18 | 199.0 | female | lunch | 35 | 3 | wed | no |
| 19 | 288.0 | female | dinner | 21 | 2 | wed | NaN |
| 20 | 120.0 | NaN | dinner | 18 | 1 | thu | yes |
| 21 | 168.0 | male | lunch | 15 | 4 | thu | yes |
| 22 | 120.0 | female | dinner | 10 | 3 | thu | NaN |
| 23 | 284.0 | male | NaN | NaN | 2 | NaN | yes |
replace() function
By using replace function you can replace a value with another value. The first parameter is the value name that you want to replace and the second parameter is that value that you want to replace with the first parameter value.
r1=df.replace("lunch","Lunch new")
print(r1)
r2=df.replace(["female","sat"],["New female","New sat"])
print(r2)
r3=df.replace(["female","sat"],"Value change")
print(r3)
Output
| total_bill | sex | time | tip | size | day | smoker | |
|---|---|---|---|---|---|---|---|
| 0 | 500.0 | male | Lunch | 30 | 2 | New sat | no |
| 1 | 648.0 | male | dinner | 35 | 3 | New sat | yes |
| 2 | 75.0 | New female | NaN | 10 | 1 | New sat | no |
| 3 | 159.0 | New female | NaN | 12 | 4 | New sat | NaN |
| 4 | 250.0 | NaN | Lunch | 40 | 3 | NaN | no |
| 5 | NaN | male | Lunch | 25 | 3 | mon | yes |
| 6 | 356.0 | male | dinner | 30 | 4 | mon | NaN |
| 7 | 99.0 | New female | NaN | 5 | 1 | mon | no |
| 8 | 150.0 | New female | dinner | 15 | 4 | mon | NaN |
| 9 | NaN | NaN | Lunch | 38 | 4 | mon | no |
| 10 | 478.0 | NaN | Lunch | 28 | 3 | NaN | yes |
| 11 | 320.0 | New female | NaN | 14 | 3 | tue | yes |
| 12 | NaN | male | Lunch | 32 | 2 | tue | NaN |
| 13 | 520.0 | male | Lunch | 33 | 3 | NaN | yes |
| 14 | 367.0 | male | dinner | 40 | 3 | NaN | yes |
| 15 | NaN | NaN | NaN | 40 | 4 | wed | yes |
| 16 | 100.0 | New female | dinner | 6 | 1 | wed | NaN |
| 17 | NaN | New female | NaN | 40 | 1 | wed | yes |
| 18 | 199.0 | NaN | Lunch | 35 | 3 | NaN | no |
| 19 | 288.0 | male | dinner | 21 | 2 | wed | NaN |
| 20 | 120.0 | New female | dinner | 18 | 1 | thu | yes |
| 21 | 168.0 | male | Lunch | 15 | 4 | NaN | yes |
| 22 | NaN | New female | dinner | 10 | 3 | thu | NaN |
| 23 | 284.0 | male | NaN | 35 | 2 | NaN | yes |
| total_bill | sex | time | tip | size | day | smoker | |
|---|---|---|---|---|---|---|---|
| 0 | 500.0 | male | Lunch new | 30 | 2 | sat | no |
| 1 | 648.0 | male | dinner | 35 | 3 | sat | yes |
| 2 | 75.0 | female | NaN | 10 | 1 | sat | no |
| 3 | 159.0 | female | NaN | 12 | 4 | sat | NaN |
| 4 | 250.0 | NaN | Lunch new | 40 | 3 | NaN | no |
| 5 | NaN | male | Lunch new | 25 | 3 | mon | yes |
| 6 | 356.0 | male | dinner | 30 | 4 | mon | NaN |
| 7 | 99.0 | female | NaN | 5 | 1 | mon | no |
| 8 | 150.0 | female | dinner | 15 | 4 | mon | NaN |
| 9 | NaN | NaN | Lunch new | 38 | 4 | mon | no |
| 10 | 478.0 | NaN | Lunch new | 28 | 3 | NaN | yes |
| 11 | 320.0 | female | NaN | 14 | 3 | tue | yes |
| 12 | NaN | male | Lunch new | 32 | 2 | tue | NaN |
| 13 | 520.0 | male | Lunch new | 33 | 3 | NaN | yes |
| 14 | 367.0 | male | dinner | 40 | 3 | NaN | yes |
| 15 | NaN | NaN | NaN | 40 | 4 | wed | yes |
| 16 | 100.0 | female | dinner | 6 | 1 | wed | NaN |
| 17 | NaN | female | NaN | 40 | 1 | wed | yes |
| 18 | 199.0 | NaN | Lunch new | 35 | 3 | NaN | no |
| 19 | 288.0 | male | dinner | 21 | 2 | wed | NaN |
| 20 | 120.0 | female | dinner | 18 | 1 | thu | yes |
| 21 | 168.0 | male | Lunch new | 15 | 4 | NaN | yes |
| 22 | NaN | female | dinner | 10 | 3 | thu | NaN |
| 23 | 284.0 | male | NaN | 35 | 2 | NaN | yes |
| total_bill | sex | time | tip | size | day | smoker | |
|---|---|---|---|---|---|---|---|
| 0 | 500.0 | male | lunch | 30 | 2 | Value change | no |
| 1 | 648.0 | male | dinner | 35 | 3 | Value change | yes |
| 2 | 75.0 | Value change | NaN | 10 | 1 | Value change | no |
| 3 | 159.0 | Value change | NaN | 12 | 4 | Value change | NaN |
| 4 | 250.0 | NaN | lunch | 40 | 3 | NaN | no |
| 5 | NaN | male | lunch | 25 | 3 | mon | yes |
| 6 | 356.0 | male | dinner | 30 | 4 | mon | NaN |
| 7 | 99.0 | Value change | NaN | 5 | 1 | mon | no |
| 8 | 150.0 | Value change | dinner | 15 | 4 | mon | NaN |
| 9 | NaN | NaN | lunch | 38 | 4 | mon | no |
| 10 | 478.0 | NaN | lunch | 28 | 3 | NaN | yes |
| 11 | 320.0 | Value change | NaN | 14 | 3 | tue | yes |
| 12 | NaN | male | lunch | 32 | 2 | tue | NaN |
| 13 | 520.0 | male | lunch | 33 | 3 | NaN | yes |
| 14 | 367.0 | male | dinner | 40 | 3 | NaN | yes |
| 15 | NaN | NaN | NaN | 40 | 4 | wed | yes |
| 16 | 100.0 | Value change | dinner | 6 | 1 | wed | NaN |
| 17 | NaN | Value change | NaN | 40 | 1 | wed | yes |
| 18 | 199.0 | NaN | lunch | 35 | 3 | NaN | no |
| 19 | 288.0 | male | dinner | 21 | 2 | wed | NaN |
| 20 | 120.0 | Value change | dinner | 18 | 1 | thu | yes |
| 21 | 168.0 | male | lunch | 15 | 4 | NaN | yes |
| 22 | NaN | Value change | dinner | 10 | 3 | thu | NaN |
| 23 | 284.0 | male | NaN | 35 | 2 | NaN | yes |
dropna() function
Dropna function is used to delete rows, columns that contain nan value. Here axis plays a very important role. By default, the axis is 0, which means that this function will work row-wise and if it gets any nan in a row then it will drop the row but if axis=1, then this function will search column wise and will drop column if gets any nan. If it doesn't pass any parameter then it will drop all those rows (if axis=1 then will drop all the columns), which have at least a single(if have more then will also drop.) nan value. If you use how=" all" then the dropna function will drop those columns or rows which have all the nan values. You can also define that, how many nan values are required to drop a row or column by using thresh parameter.
d1=df.dropna()
print(d1)
d2=df.dropna(axis=1)
print(d2)
d3=df.dropna(how="all")
print(d3)
d4=df.dropna(thresh=4,axis=0)
print(d4)
Output
| total_bill | sex | time | tip | size | day | smoker | |
|---|---|---|---|---|---|---|---|
| 0 | 500.0 | male | lunch | 30 | 2 | sat | no |
| 1 | 648.0 | male | dinner | 35 | 3 | sat | yes |
| 20 | 120.0 | female | dinner | 18 | 1 | thu | yes |
| time | tip | |
|---|---|---|
| 0 | 30 | 2 |
| 1 | 35 | 3 |
| 2 | 10 | 1 |
| 3 | 12 | 4 |
| 4 | 40 | 3 |
| 5 | 25 | 3 |
| 6 | 30 | 4 |
| 7 | 5 | 1 |
| 8 | 15 | 4 |
| 9 | 38 | 4 |
| 10 | 28 | 3 |
| 11 | 14 | 3 |
| 12 | 32 | 2 |
| 13 | 33 | 3 |
| 14 | 40 | 3 |
| 15 | 40 | 4 |
| 16 | 6 | 1 |
| 17 | 40 | 1 |
| 18 | 35 | 3 |
| 19 | 21 | 2 |
| 20 | 18 | 1 |
| 21 | 15 | 4 |
| 22 | 10 | 3 |
| 23 | 35 | 2 |
| total_bill | sex | time | tip | size | day | smoker | |
|---|---|---|---|---|---|---|---|
| 0 | 500.0 | male | lunch | 30 | 2 | sat | no |
| 1 | 648.0 | male | dinner | 35 | 3 | sat | yes |
| 2 | 75.0 | female | NaN | 10 | 1 | sat | no |
| 3 | 159.0 | female | NaN | 12 | 4 | sat | NaN |
| 4 | 250.0 | NaN | lunch | 40 | 3 | NaN | no |
| 5 | NaN | male | lunch | 25 | 3 | mon | yes |
| 6 | 356.0 | male | dinner | 30 | 4 | mon | NaN |
| 7 | 99.0 | female | NaN | 5 | 1 | mon | no |
| 8 | 150.0 | female | dinner | 15 | 4 | mon | NaN |
| 9 | NaN | NaN | lunch | 38 | 4 | mon | no |
| 10 | 478.0 | NaN | lunch | 28 | 3 | NaN | yes |
| 11 | 320.0 | female | NaN | 14 | 3 | tue | yes |
| 12 | NaN | male | lunch | 32 | 2 | tue | NaN |
| 13 | 520.0 | male | lunch | 33 | 3 | NaN | yes |
| 14 | 367.0 | male | dinner | 40 | 3 | NaN | yes |
| 15 | NaN | NaN | NaN | 40 | nan | wed | yes |
| 16 | 100.0 | female | dinner | 6 | 1 | wed | NaN |
| 17 | NaN | female | NaN | 40 | 1 | wed | yes |
| 18 | 199.0 | NaN | lunch | 35 | 3 | NaN | no |
| 19 | 288.0 | male | dinner | 21 | 2 | wed | NaN |
| 20 | 120.0 | female | dinner | 18 | 1 | thu | yes |
| 21 | 168.0 | male | lunch | 15 | 4 | NaN | yes |
| 22 | NaN | female | dinner | 10 | 3 | thu | NaN |
| 23 | 284.0 | male | NaN | 35 | 2 | NaN | yes |
| total_bill | sex | time | tip | size | day | smoker | |
|---|---|---|---|---|---|---|---|
| 0 | 500.0 | male | lunch | 30 | 2 | sat | no |
| 1 | 648.0 | male | dinner | 35 | 3 | sat | yes |
| 2 | 75.0 | female | NaN | 10 | 1 | sat | no |
| 3 | 159.0 | female | NaN | 12 | 4 | sat | NaN |
| 4 | 250.0 | NaN | lunch | 40 | 3 | NaN | no |
| 5 | NaN | male | lunch | 25 | 3 | mon | yes |
| 6 | 356.0 | male | dinner | 30 | 4 | mon | NaN |
| 7 | 99.0 | female | NaN | 5 | 1 | mon | no |
| 8 | 150.0 | female | dinner | 15 | 4 | mon | NaN |
| 9 | NaN | NaN | lunch | 38 | 4 | mon | no |
| 10 | 478.0 | NaN | lunch | 28 | 3 | NaN | yes |
| 11 | 320.0 | female | NaN | 14 | 3 | tue | yes |
| 12 | NaN | male | lunch | 32 | 2 | tue | NaN |
| 13 | 520.0 | male | lunch | 33 | 3 | NaN | yes |
| 14 | 367.0 | male | dinner | 40 | 3 | NaN | yes |
| 15 | NaN | NaN | NaN | 40 | nan | wed | yes |
| 16 | 100.0 | female | dinner | 6 | 1 | wed | NaN |
| 17 | NaN | female | NaN | 40 | 1 | wed | yes |
| 18 | 199.0 | NaN | lunch | 35 | 3 | NaN | no |
| 19 | 288.0 | male | dinner | 21 | 2 | wed | NaN |
| 20 | 120.0 | female | dinner | 18 | 1 | thu | yes |
| 21 | 168.0 | male | lunch | 15 | 4 | NaN | yes |
| 22 | NaN | female | dinner | 10 | 3 | thu | NaN |
| 23 | 284.0 | male | NaN | 35 | 2 | NaN | yes |
Fillna() function
Fillna is used to fill missing values with a specific value. If you pass method="ffill" in the fillna function then all the missing values will be filled by the previous value. If you pass method="bfill" in the fillna function then all the missing values will be filled by the next value.
f1=df.fillna("FFFFF")
print(f1)
f2=df.fillna(method="ffill")
print(f2)
f3=df.fillna(method="bfill")
print(f3) Output
| total_bill | sex | time | tip | size | day | smoker | |
|---|---|---|---|---|---|---|---|
| 0 | 500.0 | male | lunch | 30 | 2 | sat | no |
| 1 | 648.0 | male | dinner | 35 | 3 | sat | yes |
| 2 | 75.0 | female | FFFFF | 10 | 1 | sat | no |
| 3 | 159.0 | female | FFFFF | 12 | 4 | sat | FFFFF |
| 4 | 250.0 | FFFFF | lunch | 40 | 3 | FFFFF | no |
| 5 | FFFFF | male | lunch | 25 | 3 | mon | yes |
| 6 | 356.0 | male | dinner | 30 | 4 | mon | FFFFF |
| 7 | 99.0 | female | FFFFF | 5 | 1 | mon | no |
| 8 | 150.0 | female | dinner | 15 | 4 | mon | FFFFF |
| 9 | FFFFF | FFFFF | lunch | 38 | 4 | mon | no |
| 10 | 478.0 | FFFFF | lunch | 28 | 3 | FFFFF | yes |
| 11 | 320.0 | female | FFFFF | 14 | 3 | tue | yes |
| 12 | FFFFF | male | lunch | 32 | 2 | tue | FFFFF |
| 13 | 520.0 | male | lunch | 33 | 3 | FFFFF | yes |
| 14 | 367.0 | male | dinner | 40 | 3 | FFFFF | yes |
| 15 | FFFFF | FFFFF | FFFFF | 40 | 4 | wed | yes |
| 16 | 100.0 | female | dinner | 6 | 1 | wed | FFFFF |
| 17 | FFFFF | female | FFFFF | 40 | 1 | wed | yes |
| 18 | 199.0 | FFFFF | lunch | 35 | 3 | FFFFF | no |
| 19 | 288.0 | male | dinner | 21 | 2 | wed | FFFFF |
| 20 | 120.0 | female | dinner | 18 | 1 | thu | yes |
| 21 | 168.0 | male | lunch | 15 | 4 | FFFFF | yes |
| 22 | FFFFF | female | dinner | 10 | 3 | thu | FFFFF |
| 23 | 284.0 | male | FFFFF | 35 | 2 | FFFFF | yes |
| total_bill | sex | time | tip | size | day | smoker | |
|---|---|---|---|---|---|---|---|
| 0 | 500.0 | male | lunch | 30 | 2 | sat | no |
| 1 | 648.0 | male | dinner | 35 | 3 | sat | yes |
| 2 | 75.0 | female | dinner | 10 | 1 | sat | no |
| 3 | 159.0 | female | dinner | 12 | 4 | sat | no |
| 4 | 250.0 | female | lunch | 40 | 3 | sat | no |
| 5 | 250.0 | male | lunch | 25 | 3 | mon | yes |
| 6 | 356.0 | male | dinner | 30 | 4 | mon | yes |
| 7 | 99.0 | female | dinner | 5 | 1 | mon | no |
| 8 | 150.0 | female | dinner | 15 | 4 | mon | no |
| 9 | 150.0 | female | lunch | 38 | 4 | mon | no |
| 10 | 478.0 | female | lunch | 28 | 3 | mon | yes |
| 11 | 320.0 | female | NalunchN | 14 | 3 | tue | yes |
| 12 | 320.0 | male | lunch | 32 | 2 | tue | yes |
| 13 | 520.0 | male | lunch | 33 | 3 | tue | yes |
| 14 | 367.0 | male | dinner | 40 | 3 | tue | yes |
| 15 | 367.0 | male | dinner | 40 | 4 | wed | yes |
| 16 | 100.0 | female | dinner | 6 | 1 | wed | yes |
| 17 | 100.0 | female | dinner | 40 | 1 | wed | yes |
| 18 | 199.0 | female | lunch | 35 | 3 | wed | no |
| 19 | 288.0 | male | dinner | 21 | 2 | wed | no |
| 20 | 120.0 | female | dinner | 18 | 1 | thu | yes |
| 21 | 168.0 | male | lunch | 15 | 4 | thu | yes |
| 22 | 100.0 | female | dinner | 10 | 3 | thu | yes |
| 23 | 284.0 | male | dinner | 35 | 2 | thu | yes |
| total_bill | sex | time | tip | size | day | smoker | |
|---|---|---|---|---|---|---|---|
| 0 | 500.0 | male | lunch | 30 | 2 | sat | no |
| 1 | 648.0 | male | dinner | 35 | 3 | sat | yes |
| 2 | 75.0 | female | lunch | 10 | 1 | sat | no |
| 3 | 159.0 | female | lunch | 12 | 4 | sat | no |
| 4 | 250.0 | male | lunch | 40 | 3 | mon | no |
| 5 | 356.0 | male | lunch | 25 | 3 | mon | yes |
| 6 | 356.0 | male | dinner | 30 | 4 | mon | no |
| 7 | 99.0 | female | dinner | 5 | 1 | mon | no |
| 8 | 150.0 | female | dinner | 15 | 4 | mon | no |
| 9 | 478.0 | female | lunch | 38 | 4 | mon | no |
| 10 | 478.0 | female | lunch | 28 | 3 | tue | yes |
| 11 | 320.0 | female | lunch | 14 | 3 | tue | yes |
| 12 | 520 | male | lunch | 32 | 2 | tue | yes |
| 13 | 520.0 | male | lunch | 33 | 3 | wed | yes |
| 14 | 367.0 | male | dinner | 40 | 3 | wed | yes |
| 15 | 100 | female | dinner | 40 | 4 | wed | yes |
| 16 | 100.0 | female | dinner | 6 | 1 | wed | yes |
| 17 | 199 | female | lunch | 40 | 1 | wed | yes |
| 18 | 199.0 | male | lunch | 35 | 3 | wed | no |
| 19 | 288.0 | male | dinner | 21 | 2 | wed | yes |
| 20 | 120.0 | female | dinner | 18 | 1 | thu | yes |
| 21 | 168.0 | male | lunch | 15 | 4 | thu | yes |
| 22 | 284 | female | dinner | 10 | 3 | thu | yes |
| 23 | 284.0 | male | NaN | 35 | 2 | NaN | yes |
interpolate() function
This function used to fill the nan value.
new_clean=df.interpolate(method="index")
new_clean
new_clean=df.interpolate(method="polynomial",order=5)
new_clean
Clean data with random values
Here you will create random sample values and then will fill the nan cells using those random sample values. Random sample values will be created according to other values present in the selected column.
import pandas as pd
df=pd.read_csv("restaurant_bill.csv")
pd.set_option("display.max_columns",None)
print(df)
def fill_nan(dataset,column_name):
dataset[column_name]=dataset[column_name]
"""
Where the null value is present in the selected column there we are filling it with some random value.The samples are taken from the real values which are present in the selected column.After this, We will store it in a variable
"""
random_sample=dataset[column_name].dropna().sample(dataset[column_name].isnull().sum(),random_state=0)
"""
pandas need the same index to merge datasets. Now we will take those index numbers where nan is present in the selected column
"""
random_sample.index=dataset[dataset[column_name].isnull()].index
#Now we are filling the nan values by the random values
dataset.loc[dataset[column_name].isnull(),column_name]=random_sample
fill_nan(df,"tip")
df
#for showing an example we just fill tip column nan values
Output
| total_bill | sex | time | tip | size | day | smoker | |
|---|---|---|---|---|---|---|---|
| 0 | 500.0 | male | lunch | 30 | 2 | sat | no |
| 1 | 648.0 | male | dinner | 35 | 3 | sat | yes |
| 2 | 75.0 | female | NaN | 10 | 1 | sat | no |
| 3 | 159.0 | female | NaN | 12 | 4 | sat | NaN |
| 4 | 250.0 | male | lunch | 3 | 3 | NaN | no |
| 5 | 222.0 | female | lunch | 25 | 3 | mon | yes |
| 6 | 356.0 | male | dinner | 30 | 4 | mon | NaN |
| 7 | 99.0 | female | NaN | 5 | 1 | mon | no |
| 8 | 150.0 | male | dinner | 15 | 4 | mon | NaN |
| 9 | NaN | female | lunch | 38 | 4 | mon | no |
| 10 | 478.0 | female | lunch | 28 | 333 | tue | yes |
| 11 | 320.0 | nan | NaN | 14 | 3 | tue | yes |
| 12 | NaN | male | lunch | 32 | 2 | tue | NaN |
| 13 | 520.0 | male | lunch | 33 | 3 | NaN | yes |
| 14 | 367.0 | male | dinner | 30 | 3 | tue | yes |
| 15 | NaN | female | NaN | 40 | 4 | wed | yes |
| 16 | 100.0 | female | dinner | 356 | 1 | wed | NaN |
| 17 | 99.0 | Nan | NaN | 40 | 1 | wed | yes |
| 18 | 199.0 | female | lunch | 35 | 203 | wed | no |
| 19 | 288.0 | female | dinner | 21 | 102 | wed | NaN |
| 20 | 120.0 | NaN | dinner | 18 | 1 | thu | yes |
| 21 | 168.0 | male | lunch | 15 | 4 | thu | yes |
| 22 | 120.0 | female | dinner | 10 | 3 | thu | NaN |
| 23 | 284.0 | male | NaN | 26 | 2 | NaN | yes |
How to fill nan values using mean,median or mode?
import pandas as pd
df=pd.read_csv("restaurant_bill.csv")
pd.set_option("display.max_columns",None)
print(df)
def fill_nan(dataset,column_name,median):
dataset[column_name]=dataset[column_name].fillna(median)
mean_value=df.tip.mean()
median_value
fill_nan(df,"tip",median_value)
df["tip"]
Output
0 30.000000
1 35.000000
2 10.000000
3 12.000000
4 25.818182
5 25.000000
6 30.000000
7 5.000000
8 25.818182
9 38.000000
10 28.000000
11 14.000000
12 32.000000
13 33.000000
14 25.818182
15 40.000000
16 6.000000
17 40.000000
18 35.000000
19 21.000000
20 18.000000
21 25.818182
Name: tip, dtype: float64
Let's see a advance technique to handle the missing values.
Let's see the dataset
import pandas as np
df=pd.read_csv("practice2.csv")
print(df)
Output
| total_bill | sex | time | tip | size | day | smoker | |
|---|---|---|---|---|---|---|---|
| 0 | 500.0 | male | lunch | 30 | 2 | sat | no |
| 1 | 648.0 | male | dinner | 35 | 3 | sat | yes |
| 2 | 75.0 | female | dinner | 10 | 1 | sat | no |
| 3 | 159.0 | female | lunch | 12 | 4 | sat | NaN |
| 4 | 250.0 | male | lunch | 3 | 3 | NaN | no |
| 5 | 222.0 | female | lunch | 25 | 3 | mon | yes |
| 6 | 356.0 | male | dinner | 30 | 4 | mon | NaN |
| 7 | 99.0 | female | dinner | 5 | 1 | mon | no |
| 8 | 150.0 | male | dinner | NaN | 4 | mon | NaN |
| 9 | NaN | female | lunch | 38 | 4 | mon | no |
| 10 | 478.0 | female | lunch | 28 | 3 | tue | yes |
| 11 | 320.0 | nan | dinner | 14 | 3 | tue | yes |
| 12 | NaN | male | lunch | 32 | 2 | tue | NaN |
| 13 | 520.0 | male | lunch | 33 | 3 | NaN | yes |
| 14 | 367.0 | male | dinner | NaN | 3 | tue | yes |
| 15 | NaN | female | dinner | 40 | 4 | wed | yes |
| 16 | 100.0 | female | dinner | 6 | 1 | wed | NaN |
| 17 | 99.0 | Nan | lunch | 40 | 1 | wed | yes |
| 18 | 199.0 | male | lunch | 35 | 3 | wed | no |
| 19 | 288.0 | female | dinner | 21 | 2 | wed | NaN |
| 20 | 120.0 | NaN | dinner | 18 | 1 | thu | yes |
| 21 | 168.0 | male | lunch | 15 | 4 | thu | yes |
| 22 | 120.0 | female | dinner | 10 | 3 | thu | NaN |
| 23 | 284.0 | male | dinner | NaN | 2 | NaN | yes |
step 1:
Checking for how many missing values are present in each column.
df1=df.isnull().sum()
print(df1)
Output
total_bill 6
sex 5
time 0
tip 0
size 0
day 7
smoker 7
dtype: int64
Step 2:
Checking for how many missing values present in the dataset
df2=df.isnull().sum()
print(df2)
Output
25
Step 3:
Filling missing values of categorical column
Here at first print all the categorical columns names
categorical_column=df.select_dtypes(include="object").columns
print(categorical_column)
Output
Index(['sex', 'time', 'day', 'smoker'], dtype='object')
Step 4:
Let's print all the categorical columns
categorical_column=df.select_dtypes(include="object")
categorical_column.head(5)
Output
| sex | time | day | smoker | |
|---|---|---|---|---|
| 0 | male | lunch | sat | no |
| 1 | male | dinner | sat | yes |
| 2 | female | dinner | sat | no |
| 3 | female | lunch | sat | NaN |
| 4 | NaN | lunch | NaN | no |
Step 5:
Let's see the percentage of missing values contained by each categorical column.
df3=categorical_column.isnull().sum()/df.shape[0]*100
print(df3)
Output
sex 12.500000
day 16.666667
smoker 29.166667
dtype: float64
Step 6
If you have a lot of missing values present in a column then remove that column, because it will not matter which way you fill that column, it will always cause bad accuracy. What you have to do is that, you have to select a boundary for the missing value contain by each column, if a column has more missing values than the boundary, then remove that column. You already printed which column is containing how much missing values in percentage. So now print those columns names that contain more than 17 percentage missing values.
df4=df3[df3>17].keys()
df4
Output
Index([ 'smoker'], dtype='object')
Step 7:
Now drop those columns which contain missing values more than the boundary. Here boundary is 17%. You already have those columns' names.
categorical_column.drop(columns=df4,axis=1)
print(categorical_column.head())
Output
| sex | day | |
|---|---|---|
| 0 | male | sat |
| 1 | male | sat |
| 2 | female | sat |
| 3 | female | sat |
| 4 | NaN | NaN |
Step 8:
Printing unique value for time column because to filter data for categorical columns you will use it.
df10=df.sex.unique()
print(df10)
Output
['lunch' 'dinner']
Step 9:
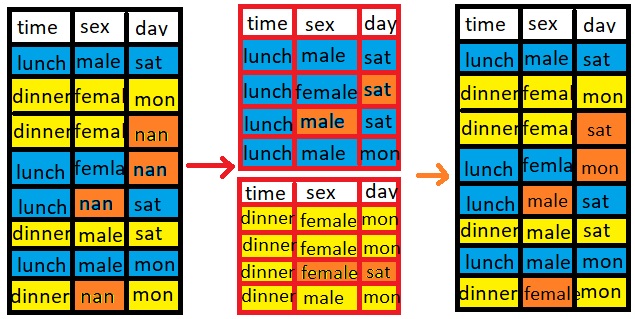
How we will fill the missing values?
For example lunch present 5 times in the time column and dinner present 4 times. Now for 5 times lunch, there
5 values should be present in sex column . But suppose only three values are present and other two values are
missing. Now we will take mode of those 3 values of sex column which have lunch value in the time column and
with that mode value we will fill other 2 missing values present in sex column which time column value is
lunch.
For dinner we will do the same.
So we can say that for lunch we will find mode value in sex and day column and will fill those two missing
values and for dinner we will find mode value of sex and day column and will fill the missing values.
dinneR=df.time=="dinner"
#------------------------------------------
#--------------------------------------------
df11=df.loc[luncH,["day","sex"]]
df12=df.loc[luncH,["day","sex"]]
#-------------------------------------------------
#--------------------------------------------------
df13=df11[["day","sex"]].mode()
df14=df12[["day","sex"]].mode()
df15=df12["sex"].fillna(df14.loc[0][1])
df16=df11["sex"].fillna(df13.loc[0][1])
temp1=pd.concat([df15,df16],ignore_index=True)
df17=df12["day"].fillna(df14.loc[0][0])
df18=df11["day"].fillna(df13.loc[0][0])
temp2=pd.concat([df17,df18],ignore_index=True)
categorical_data=pd.merge(temp1,temp2,right_index=True,left_index=True)
print(categorical_data)
Output
| sex | day | |
|---|---|---|
| 0 | male | sat |
| 1 | male | sat |
| 2 | female | sat |
| 3 | female | sat |
| 4 | male | mon |
| 5 | female | mon |
| 6 | male | mon |
| 7 | female | mon |
| 8 | male | mon |
| 9 | female | mon |
| 10 | female | tue |
| 11 | male | tue |
| 12 | male | tue |
| 13 | male | mon |
| 14 | male | tue |
| 15 | female | wed |
| 16 | female | wed |
| 17 | male | wed |
| 18 | female | wed |
| 19 | female | wed |
| 20 | male | thu |
| 21 | male | thu |
| 22 | female | thu |
| 23 | male | mon |
Step 10:
Filling numeric missing values of columns
Let's print all the numeric columns name.
numerical_column=df.select_dtypes(include=["int64","float64"]).columns
print(numerical_column)
Output
Index(['total_bill', 'tip', 'size'], dtype='object')
Step 11:
Let's print all the numeric columns
numerical_column=df.select_dtypes(include=["int64","float64"])
print(numerical_column.head())
Output
| total_bill | tip | size | |
|---|---|---|---|
| 0 | 500.0 | 30 | 2 |
| 1 | 648.0 | 35 | 3 |
| 2 | 75.0 | 10 | 1 |
| 3 | 159.0 | 12 | 4 |
| 4 | 250.0 | NaN | 3 |
Step 12:
Let's see how much missing value contains by each numeric column in percentage.
df7=numerical_column.isnull().sum()/df.shape[0]*100
df7
Output
total_bill 13.636364
tip 18.181818
size 0.000000
dtype: float64
Step 13:
Same thing what we did while categorical column in step 6.
df8=df7[df7>17].keys()
print(df8)
Output
Index(['tip'], dtype='object')
Step 14:
Drop those columns which contain missing values more than the boundary. You already have those column names.
df9=df.drop(columns=df8,axis=1)
print(df9.head())
Output
| total_bill | size | |
|---|---|---|
| 0 | 500.0 | 2 |
| 1 | 648.0 | 3 |
| 2 | 75.0 | 1 |
| 3 | 159.0 | 4 |
| 4 | 250.0 | 3 |
Step 15:
Printing unique value for time column because to filter data for numerical columns you will use it.
df10=df.time.unique()
print(df10)
Output
['lunch' 'dinner']
Step 16:
Female=df.sex=="female"
Male=df.sex=="male"
#------------------------------------------
#--------------------------------------------
df11=df.loc[Female,["size","total_bill"]]
df12=df.loc[Male,["size","total_bill"]]
df13=df11.fillna(df11.mean())
df14=df12.fillna(df12.mean())
new_numerical_column=pd.concat([df13,df14],ignore_index=False)
new_numerical_column.sort_index()
Output
| total_bill | size | |
|---|---|---|
| 0 | 500.0 | 2 |
| 1 | 648.0 | 3 |
| 2 | 75.0 | 1 |
| 3 | 159.0 | 4 |
| 4 | 250.0 | 3 |
| 5 | 222.0 | 3 |
| 6 | 356.0 | 4 |
| 7 | 99.0 | 1 |
| 8 | 150.0 | 4 |
| 9 | 363.2 | 4 |
| 10 | 478.0 | 3 |
| 11 | 320.0 | 3 |
| 12 | 363.2 | 2 |
| 13 | 520.0 | 3 |
| 14 | 367.0 | 3 |
| 15 | 363.2 | 4 |
| 16 | 100.0 | 1 |
| 17 | 99.0 | 1 |
| 18 | 199.0 | 3 |
| 19 | 288.0 | 2 |
| 20 | 120.0 | 1 |
| 21 | 168.0 | 4 |
| 22 | 120.0 | 3 |
| 23 | 284.0 | 2 |
Step 17
Now merge categorical and numerical recently cleaned columns.
catago_and_numeric=pd.merge(categorical_data,new_numerical_column,right_index=True,left_index=True)
Output
| sex | day | total_bill | size | |
|---|---|---|---|---|
| 0 | male | sat | 500.0 | 2 |
| 1 | male | sat | 648.0 | 3 |
| 2 | female | sat | 75.0 | 1 |
| 3 | female | sat | 159.0 | 4 |
| 4 | male | mon | 250.0 | 3 |
| 5 | female | mon | 222.0 | 3 |
| 6 | male | mon | 356.0 | 4 |
| 7 | female | mon | 99.0 | 1 |
| 8 | male | mon | 150.0 | 4 |
| 9 | female | mon | 363.2 | 4 |
| 10 | female | tue | 478.0 | 3 |
| 11 | male | tue | 320.0 | 3 |
| 12 | male | tue | 363.2 | 2 |
| 13 | male | mon | 520.0 | 3 |
| 14 | male | tue | 367.0 | 3 |
| 15 | female | wed | 363.2 | 4 |
| 16 | female | wed | 100.0 | 1 |
| 17 | male | wed | 99.0 | 1 |
| 18 | female | wed | 199.0 | 3 |
| 19 | female | wed | 288.0 | 2 |
| 20 | male | thu | 120.0 | 3 |
| 21 | male | thu | 168.0 | 4 |
| 22 | female | thu | 120.0 | 3 |
| 23 | male | mon | 284.0 | 2 |
Step 18
Now merge categorical and numerical recently merged data with remaining columns of the dataset which we doesn't clean to get the final clean dataset.
remaing_cols=df[["smoker","time","tip"]]
fdt=pd.merge(catago_and_numeric,remaing_cols,right_index=True,left_index=True)
fdt
Output